Value at Risk
Contents
Value at Risk#
Session objectives:
Compute Value at Risk (VaR)
Empirical VaR and Expected Shortfall on empirical data
Parametric VaR using the Normal distribution assumption
Simulate stock/index level using Geometric Brownian Motion (GBM)
Simulated VaR
Backtesting a VaR
Compute expected Shortfall
1. Import packages (matplotlib, pandas, os,…) and data#
1.1 Import packages (matplotlib, pandas, os,…)#
c’est quoi un import relatif
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import os
import statsmodels.api as sm
from scipy.stats import norm
# Remark : import all vs relative import
import math
math.sqrt(9)
3.0
from math import sqrt
sqrt(9)
3.0
1.2. Set your working directory using os.chldr (same as in the previous exercice on Volatility)#
#os.chdir(r'C:\Users\cdes1\OneDrive - ICHEC\Documents\Cours\PortfolioManagement')
os.chdir(r'/Users/christophe/OneDrive - ICHEC/Documents/Cours/PortfolioManagement')
1.3. Import and check the data#
Import the excel File
sp500_daily.xlsx. Keep data from 2010 onwards.Verify your data by showing the first five rows and the last five rows (as in the previous exercice on Volatility)
df = pd.read_excel('data/sp500_daily.xlsx', index_col=0, parse_dates=True)
df = df.loc['2010-01-01':]
df.head()
| CLOSE | |
|---|---|
| Date | |
| 2010-01-04 | 1132.9855 |
| 2010-01-05 | 1136.5234 |
| 2010-01-06 | 1137.1378 |
| 2010-01-07 | 1141.6949 |
| 2010-01-08 | 1144.9831 |
df.tail()
| CLOSE | |
|---|---|
| Date | |
| 2022-12-23 | 3844.8150 |
| 2022-12-27 | 3829.2493 |
| 2022-12-28 | 3783.2219 |
| 2022-12-29 | 3849.2832 |
| 2022-12-30 | 3839.4966 |
1.4. Compute daily log-returns in ndarray (numpy) rets to compute empirical VaR#
prices = df['CLOSE']
rets = np.log(prices / prices.shift(1))[1:]
print(rets[:10])
Date
2010-01-05 0.003118
2010-01-06 0.000540
2010-01-07 0.004000
2010-01-08 0.002876
2010-01-11 0.001742
2010-01-12 -0.009424
2010-01-13 0.008287
2010-01-14 0.002428
2010-01-15 -0.010884
2010-01-19 0.012425
Name: CLOSE, dtype: float64
Compute the number of observations (length of vector
rets)
n = len(rets)
n
3271
2. Empirical VaR#
Compute the 95% and the 99% VaR of the empirical distribustion
Use the 5th and the 1st percentile of the return distribution using the function
np.percentile([returns],q=[percentile]).The percentile should be a number between 0 and 100.
Note: the percentile 0 corresponds to the minimum, check the result with the function (
np.min) of the distribution.Note: the percentile 100 corresponds to the maximum, check the result with the function (
np.max) of the distribution.
p5 = np.percentile(rets, q=5)
print("Var(5%):", f"{p5:.4%}")
Var(5%): -1.7348%
p1=np.percentile(rets,q=1)
print("VaR(1%):",f"{p1:.4%}")
VaR(1%): -3.2953%
np.percentile(rets, q=0)
-0.12765113378213216
np.min(rets)
-0.12765113378213216
np.percentile(rets, q=100)
0.0896853763515508
np.max(rets)
0.0896853763515508
np.maximum(4, 5)
5
df.loc['2020-03-01':'2020-04-30'].plot()
<AxesSubplot:xlabel='Date'>
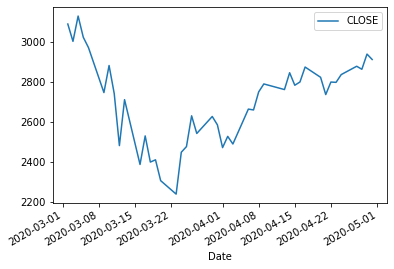
Identify when the maximum return happened
rets[rets==np.max(rets)]
Date
2020-03-24 0.089685
Name: CLOSE, dtype: float64
3. Represent the empirical distribution and check for Normality (To use parametric VaR)#
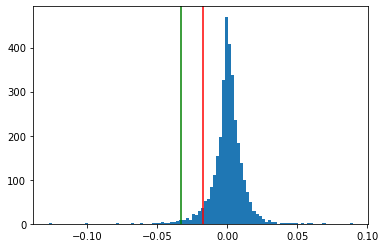
3.1. Represent the histogram of the return distribution#
Add the two estimated emprical VaR (95% and 99%) (with 2 vertical bars)
num_bins = 100
plt.hist(rets, bins=num_bins)
plt.axvline(x=p5, color='red')
plt.axvline(x=p1, color='green');
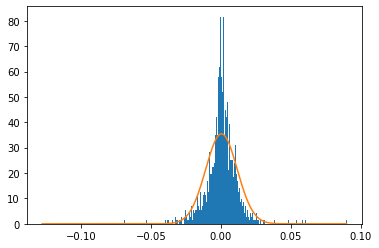
3.2. Compare the empirical ditribution with a Normal distrubution by adding a normal curve on the graph#
import scipy.stats as scs
Note : Function equivalent to =NORM.DIST(x;0;1;TRUE) in Excel
scs.norm.cdf(-1.96)
0.024997895148220435
Add a normal curve on the graph and observe the relative “Normality” of the empirical data
num_bins = 1000
plt.hist(rets, bins=num_bins, density=True);
x = np.linspace(np.min(rets), np.max(rets), 1000)
r = np.mean(rets)
pdf = scs.norm.pdf(x, loc=r, scale=np.std(rets))
#print(pdf)
plt.plot(x, pdf);
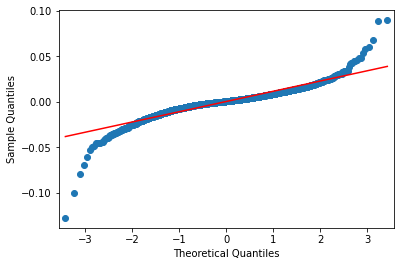
Use the qqplot to observe the compare the tails of the empiical distribution wth the tails of the Normal distribution
sm.qqplot(rets, line='s');
3.3. Test for normality#
Use the Jarque Bera test and interpret the result
\(H_0\): Data are normally distributed
\(H_1\): Data are not normally distributed
from statsmodels.stats.stattools import jarque_bera
jarque_bera(rets)
(24046.491072008783, 0.0, -0.7337810849301992, 16.201526948721195)
from scipy.stats import jarque_bera
jarque_bera(rets)
Jarque_beraResult(statistic=24046.49107200881, pvalue=0.0)
We can verify the the Jarque Bera test by coding the test step by step
Compute S
Compute K
Compute JB
from scipy.stats import skew, kurtosis
print(skew(rets))
print(kurtosis(rets))
-0.7337810849301992
13.201526948721195
m3 = np.sum((rets - np.mean(rets))**3) / n
m2 = np.sum((rets - np.mean(rets))**2) / n
Sk = m3 / m2**(3/2)
Sk
-0.7337810849301989
m4 = np.sum((rets - np.mean(rets))**4) / n
Ku = m4 / m2**2
Ku - 3
13.201526948721195
JB = n / 6 * (Sk**2 + ((Ku-3)**2)/4)
JB
24046.491072008783
Another normality test : Shapiro Wilk test#
The Shapiro-Wilk test tests the null hypothesis that the data was drawn from a normal distribution.
https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.shapiro.html
from scipy.stats import shapiro
shapiro(rets)
ShapiroResult(statistic=0.8889527916908264, pvalue=1.5694542800437951e-43)
A test for any distribution : Anderson test#
The Anderson-Darling test tests the null hypothesis that a sample is drawn from a population that follows a particular distribution. For the Anderson-Darling test, the critical values depend on which distribution is being tested against. This function works for normal, exponential, logistic, or Gumbel (Extreme Value Type I) distributions.
https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.anderson.html
from scipy.stats import anderson
anderson(rets)
AndersonResult(statistic=68.0861414366509, critical_values=array([0.575, 0.655, 0.786, 0.917, 1.091]), significance_level=array([15. , 10. , 5. , 2.5, 1. ]))
4. Compute parametric VaR (with a Normal distribution assumption)#
4.1. Compute the average daily return#
mu = np.average(rets)
print(mu)
0.0003731229227067712
4.2. Compute the daily standard deviation#
sigma = np.std(rets, ddof=1)
print(sigma)
0.011254626546227362
4.3. Compute parametric VaR#
Note : Equivalent to =NORM.INV(x;0;1) in Excel
print('99%', norm.ppf(0.01))
print('97,5%', norm.ppf(0.025))
print('95%', norm.ppf(0.05))
99% -2.3263478740408408
97,5% -1.9599639845400545
95% -1.6448536269514729
Z = norm.ppf(0.05)
mu + Z * sigma
-0.018139090371839632
p5
-0.017347574787051057
Z = norm.ppf(0.01)
mu + Z * sigma
-0.025809053616232862
p1
-0.03295346538723258
5. Simulate a data distribution with the Geometric brownian motion (GBM) to compute a VaR by Monte Carlo simulation#
df.tail()
| CLOSE | |
|---|---|
| Date | |
| 2022-12-23 | 3844.8150 |
| 2022-12-27 | 3829.2493 |
| 2022-12-28 | 3783.2219 |
| 2022-12-29 | 3849.2832 |
| 2022-12-30 | 3839.4966 |
5.1. Simulate a data distribution the next day using the GBM#
Edit (23/04/23):
T=1/252 has been replaced with T=1
r is the mean value of daily return and sigma is the standard deviation of the returns (but not annualized).
np.random.seed(0)
S0 = df.iloc[-1]['CLOSE']
r = np.mean(rets)
sigma = np.std(rets)
T = 1
numSim = 10000
ST1 = S0 * np.exp((r - 0.5 * sigma**2) * T +
np.random.standard_normal(numSim) * sigma * np.sqrt(T))
plt.hist(ST1, bins=50);
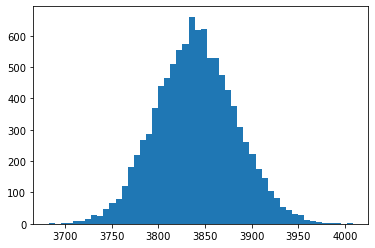
5.2. Compute the 99% VaR#
rets = np.log(ST1 / S0)
plt.hist(rets, bins=1000);
print(np.percentile(rets, q=1))
-0.02563021860323059
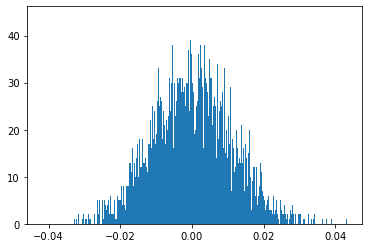
5.3. Simulate a single return trajectory on one year and a distribution in one year#
Edit (23/04/23):
mu is the mean value of daily return multiplied by 252. Therefore it represents the annual mean.
sigma is the annualized standard deviation of the returns.
np.random.seed(1)
T = 252
mu = np.mean(rets)*T # new
sigma = np.std(rets)*sqrt(T) # new
S = np.zeros(T+1)
S[0] = S0
dt = 1/252
for t in range(T):
S[t+1] = S[t] * np.exp((mu-sigma**2/2)*dt + sigma * np.random.normal(0, 1) * np.sqrt(dt))
plt.plot(S);
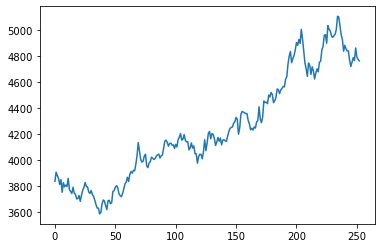
numSim = 10000
T = 252
dt = 1 / T
mu = np.mean(rets)*T # new
sigma = np.std(rets)*np.sqrt(T) # new
S = np.zeros((T+1, numSim))
S[0] = S0
for t in range(1, T+1):
S[t] = S[t-1] * np.exp((mu - sigma**2/2) * dt +
sigma * np.sqrt(dt) * np.random.standard_normal(numSim))
plt.hist(S[-1], bins=50);
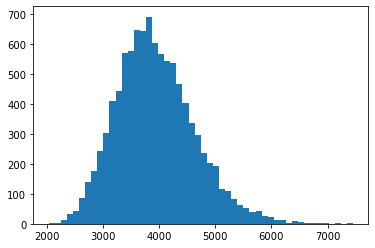
S
array([[3839.4966 , 3839.4966 , 3839.4966 , ..., 3839.4966 ,
3839.4966 , 3839.4966 ],
[3722.30340007, 3923.22264717, 3855.31569663, ..., 3861.11194267,
3873.85568493, 3760.96120326],
[3748.39880064, 3937.33272466, 3908.94398872, ..., 3861.53992374,
3859.69168867, 3717.22701904],
...,
[2925.63745469, 3947.4365368 , 3051.6340238 , ..., 3729.83556295,
4826.42577662, 3981.43817732],
[2902.6029258 , 4000.91410254, 3031.00608792, ..., 3727.39843353,
4839.9391242 , 4008.86906069],
[2899.37229201, 3960.90670635, 3034.68939292, ..., 3717.6130952 ,
4807.63832887, 4061.56943795]])
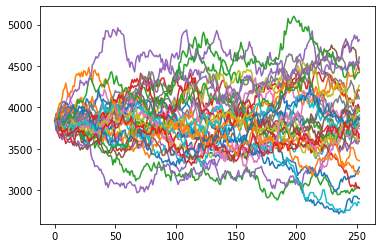
plt.plot(S[:,:35]);
6. VaR Backtesting and the binomial distribution#
LOI.BINOMIALE(1 ; 250 ; 0.01 ; faux) = 0.205 (probability)
LOI.BINOMIALE(1 ; 250 ; 0,01 ; vrai) = 0.286 (cumulative probability)
from scipy.stats import binom
x = 1 # x succes
n = 250 # n essais
p = 0.01 # proba
binom.pmf(x, n, p) # pmf = probability mass function
0.20469322263177145
from math import factorial
def combi(n, x):
c = factorial(n)/(factorial(x)*factorial(n-x))
return c
def binomial(x, n, p):
return combi(n, x) * p**x * (1-p)**(n-x)
binomial(x, n, p)
0.2046932226317709
x = 1
n = 250
p = 0.01
binom.cdf(x, n, p)
0.28575173879395216
def Binomial(x, n, p):
b = 0
for j in range(x+1):
b += binomial(j, n, p)
return b
Binomial(x, n, p)
0.28575173879395216
LOI.BINOMIALE(4;250;0,01;faux) = 0,134 (probabilité)
LOI.BINOMIALE(4;250;0,01;vrai) = 0,892 (probabilité cumulée 0,1,2,3 et 4)
x = 4
n = 250
p = 0.01
print(binom.pmf(x, n, p))
print(binom.cdf(x, n, p))
0.13407092913874208
0.8921876269036252
x = 5
n = 250
p = 0.01
print(binom.pmf(x, n, p))
print(binom.cdf(x, n, p))
0.06662918902652627
0.9588168159301517
7. Conditional VaR / Expected Shortfall#
7.1. Keep the 5th data percentile and count the observations#
rets = np.log(df / df.shift(1))[1:]
rets = np.array(rets)
p5 = np.percentile(rets, q=5)
p5
-0.017347574787051057
low_returns = rets[rets < p5]
len(low_returns) / len(rets)
0.05013757260776521
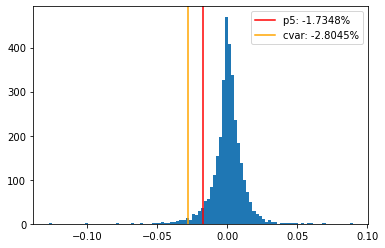
7.2. Compute the average of the 5th tail data and show the ES on the distribution graph#
cvar = np.mean(low_returns)
cvar
-0.028044792302960128
plt.hist(rets, bins=100)
plt.axvline(p5, color='red', label='p5: '+f"{p5:.4%}")
plt.axvline(cvar, color='orange', label='cvar: '+f"{cvar:.4%}")
plt.legend();
References#
https://www.statsmodels.org/dev/_modules/statsmodels/stats/stattools.html
https://github.com/yhilpisch/py4fi2nd/
https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.shapiro.html
https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.anderson.html

